A chromosome-level genome assembly of the Chinese herbal medicine Chelidonium majus
Sample collection
All specimens were collected following the guidelines of the Earth Biogenome Project ( Fresh leaves and roots of Chelidonium majus were collected from fields (30.86°N, 120.19°E) in Huzhou, Zhejiang, China in March 2024. Samples were immediately stored at −80°C until DNA extraction. Each sample was associated with a properly preserved voucher specimen, deposited in Zhejiang Institute of Freshwater Fisheries under catalog number (ZIFF-CM-001 and ZIFF-CM-002).
DNA/RNA extraction
The leaves samples were used for DNA isolation by standard CTAB method. First, samples were lysed in 1000 μL of CTAB buffer and supplemented with 20 μL lysozyme, followed by incubation at 65 °C for 2-3 hours with periodic mixing. After centrifugation, 950 μL of supernatant was extracted with an equal volume of phenol: chloroform: isoamyl alcohol (25:24:1), followed by a second extraction using chloroform: isoamyl alcohol (24:1). The DNA was then precipitated by adding 3/4 volume isopropanol and incubating at −20 °C. Subsequent steps included centrifugation, washing the pellet twice with 75% ethanol, and air-drying the DNA under sterile conditions. The purified DNA was resuspended in 51 μL ddH2O, with optional heating at 55–60 °C to facilitate dissolution. Finally, residual RNA was removed by adding 1 μL RNase A and incubating at 37 °C for 15 minutes. Both leaves and roots were subjected to RNA isolation using Trizol reagent (Invitrogen, CA, USA). The quantity of DNA and RNA were examined by a Qubit 3.0 Fluorometer (Thermo Fisher Scientific, Waltham, USA) and a Bioanalyzer 2100 system (Agilent Technologies, CA, USA), respectively. The results showed that the concentration of DNA was 232 ng/μL, with the A260/A280 and A260/A230 values of 1.80 and 2.10, respectively. The concentration of RNA was 160 ng/μL, with the RIN value of 6.9. The quality of extracted DNA and RNA were evaluated using agarose gel electrophoresis and NanoDrop 2000 spectrophotometer (NanoDrop Technologies, Wilmington, USA). DNA and RNA concentrations were determined to be 253.22 ng/μL and 168.40 ng/μL, respectively.
Library preparing and sequencing
For the short reads sequencing, the qualified DNA sample was randomly fragmented using the Covaris ultrasonic disruptor, followed by library generation with an insert size of 350 bp. For Hi-C sequencing, Hi-C libraries were prepared and constructed according to the previously described methods11. After quality inspection, all the constructed libraries were subjected to 150 bp paired-end (PE) sequencing on the Illumina NovaSeq 6000 platform (Illumina, CA, USA). For PacBio sequencing, a SMRTbell library was constructed using SMRTbell Express Template Prep Kit 2.0 (Pacific Biosciences, CA, USA). AMPure PB Beads were used to concentrate and purify the library. The constructed library was then sequenced on the PacBio Sequel II platform. For transcriptome sequencing, the TruSeqTM RNA Sample Preparation Kit (Illumina, CA, USA) was used to construct RNA-seq transcriptome libraries and followed by sequencing on the Illumina NovaSeq 6000 platform. Besides, Iso-Seq Express 2.0 Kit (Pacific Biosciences, CA, USA) and Kinnex full-length RNA Kit (Pacific Biosciences, CA, USA) were used to synthesis cDNA and construct library, respectively. The library was then subjected to sequencing with the PacBio Sequel II platform. In summary, 68.14 Gb short reads, 37.40 Gb PacBio reads, 114.28 Hi-C reads, and 47.11 RNA-seq reads of Chelidonium majus were generated in this study (Table 1).
Genome size and heterozygosity estimation
Adaptors and low-quality reads were removed from the raw data using fastp (v0.21.0)12. The clean data was employed for genome size estimation. K-mer analysis was conducted using the software Jellyfish (v2.2.7)13. K-mer 17 was used to conduct survey analysis. The results showed that the genome size of C. majus was estimated to be 1,118.6 Mb, with the heterozygous ratio of 1.07% (Table 2).
De novo Genome assembly and chromosome construction
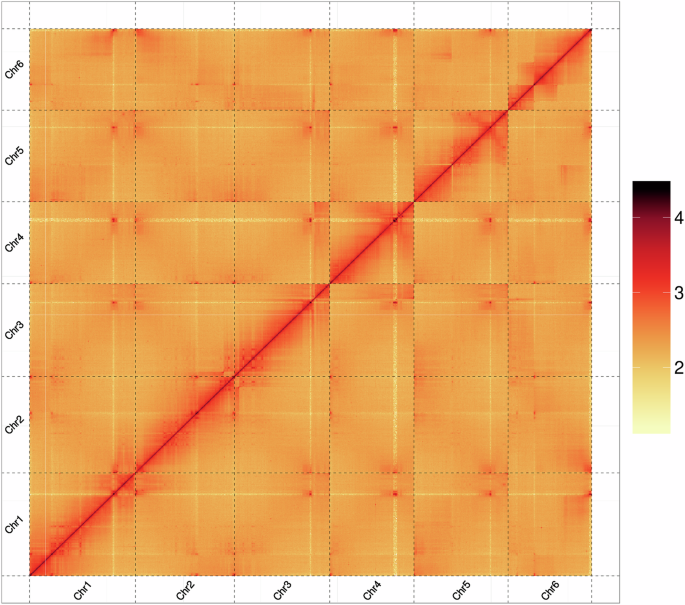
For the de novo genome assembly, a hybrid strategy was adopted, combining the both clean PacBio HiFi reads and Illumina Hi-C reads. First, use the CCS ( parameters: min-rq = 0.99) to perform quality control on the 37.4 Gb raw HiFi sequencing data. The resulting high-fidelity reads were subsequently assembled into contigs using the Hifiasm (v0.19.8)14 with default parameters. To achieve chromosome-level scaffolding, the contig assembly was integrated with the sequenced 114.28 Gb Hi-C data through the ALLHiC pipeline15, including five steps: pruning, partition, rescue, optimization, building. Final manual refinement was performed using Juicebox (v1.11.08)16. The heatmap of both intra- and inter-chromosomal interactions was visualized (Fig. 1). A 918,794,832 bp (91.21%) of sequences were successfully anchored onto 6 pseudo-chromosomes. Estimated genome information in the C-value database at Kew ( showed that the estimated genome size of 1.107 Gb and chromosome number of 2n = 2x = 12, which provided independent support for the assembly in this study. Finally, the assembled genome amounted to 1.06 Gb, comprising 1,520 contigs, with an N50 of 106.65 Mb (Table 3). The circos plot of C. majus genome was shown in Fig. 2.

Hi-C interaction analysis.

Circos plot of Chelidonium majus genome illustrating from outside to inside: (a) chromosome length, (b) gene density, (c) TE density, (d) GC content, (e) collinearity.
Repetitive sequence annotation
Repetitive sequence annotation was performed using a combination of homology-based sequence alignment and de novo prediction approaches. For the homology-based sequence alignment, RepeatMasker (v4.1.6)17 was employed to search against the Repbase TE library18 to identify sequences similar to known repetitive elements. For the de novo prediction, a de novo repetitive sequence library was first constructed using RepeatModeler ( followed by de novo repeat prediction. Finally, a total of 697,778,264 bp of repetitive sequences were identified in the assemble genome of C. majus (Table 4), including short interspersed nuclear element (SINE, 1.07%), short interspersed nuclear element (LINE, 5.92%), long terminal repeat (LTR, 45.08%), DNA transposon (15.79%), and unknown element (1.00%), which occupied 69.27% of the genome.
Gene structure prediction
For the gene structure prediction, a comprehensive approach combining de novo, homology-based, and transcriptome-based methods was used to predict genes within the assembled genome. For homology-based prediction, protein sequences from Arabidopsis thaliana (Atha) (Col-PEK1.5), Macleaya cordat (Mcor) (GCA 002174775.1), and Papaver somniferum (Psom) (GCF 003573695.1) were collected for mapping onto the C. majus genome using TBLASTN19 with an e-value ≤ 10−5. For the de novo prediction, Augustus (v3.5.0)20 and SNAP ( were used to predict gene coding regions with default parameters. For transcriptome-based gene prediction, Trinity(v2.8)21 was first used to perform transcriptome assembly, followed by predicting the gene structure by PASA(v2.5.2)22. EVidenceModeler(EVM)v1.1.1( was employed to merge the gene sets predicted by the various methods into a non-redundant and more comprehensive gene set. Subsequently, the PASA pipeline (http://pasa.sourceforge.net)23 was employed to refine the EVM annotations by incorporating transcriptome assembly data to produce the final gene set. A total of 25,203 protein-coding genes were identified. The average CDS length was 1,258.59 bp. The average exon number per gene was 5.11 with an average exon length of 246.34 bp and average intron length of 596.13 bp (Table 5). AGAT Tool kit ( also was used to assess this genome. The result showed that the number of genes containing only 3’UTR is 808, the number of genes containing only 5’UTR is 238, and the number of genes containing both 3’UTR and 5’UTR is 14,369. The number of single exon genes was 4766.
Gene function prediction
For the gene function prediction, the protein sequences were aligned against known protein libraries including National Center for Biotechnology Information (NCBI) Non-Redundant (NR), Swiss-Prot24, InterPro25, and Pfam26 databases using BLAST19 with an e-value ≤ 10−5 (access time: July 10, 2024). Blast2GO(v6.0)27 was employed to annotate functions and pathways based on the Gene ontology (GO)28 and Kyoto Encyclopedia of Genes and Genomes (KEGG)29 databases (access time: July 10, 2024). A total of 24,749 protein-coding genes were successfully predicted (Table 6 and Fig. 3).

Venn diagram of function annotations from different databases.
Non-coding RNA annotation
For the non-coding RNA annotation, tRNAscan-SE30 was used for the tRNA prediction and ribosomal RNAs (rRNAs) were identified by BLAST. miRNA and snRNA were predicted by using Infernal (v1.1)31 against the Rfam database32. The results of non-coding RNA annotation were shown in Table 7.
link







